Abstract
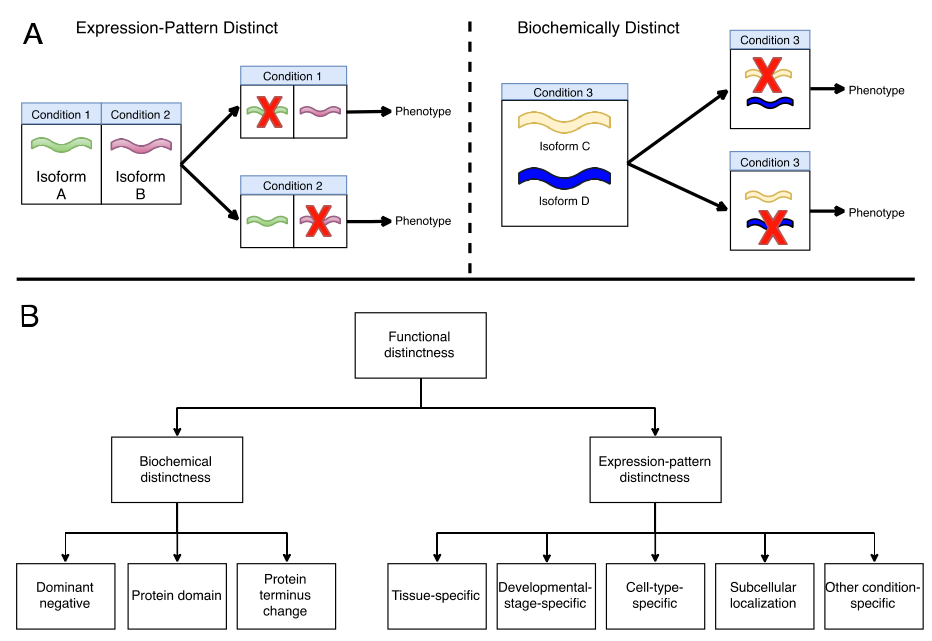
- Classify distinct functions of isoforms
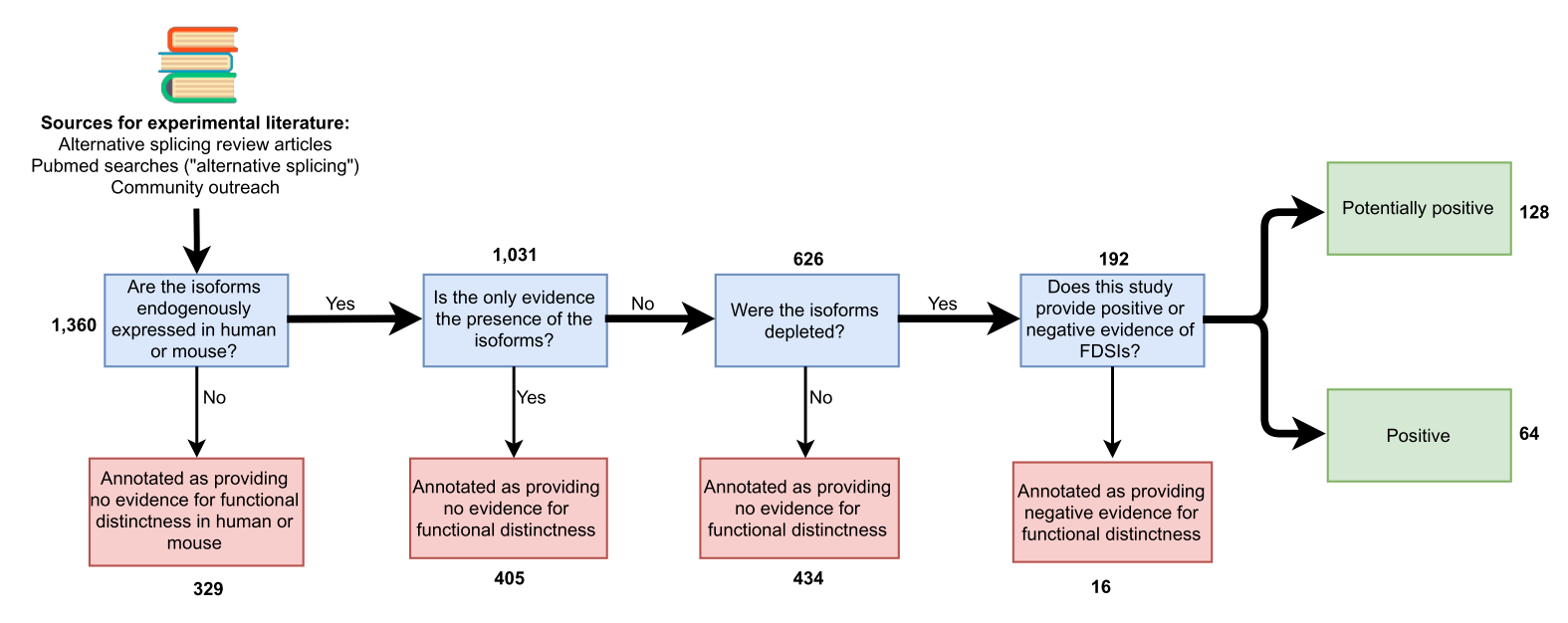
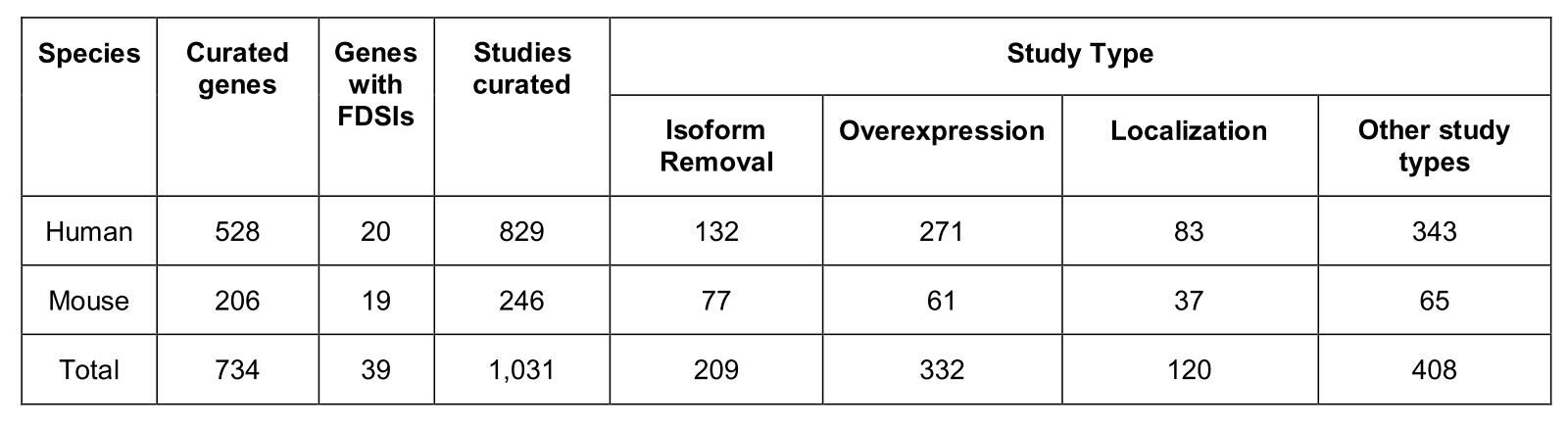
- Literature-based analysis of experimental evidence for functionally distinct splice isoforms (FDSIs) for over 700 human & mouse genes
- Result: fewer than 10% of genes have solid experimental evidences of FDSIs.
Main
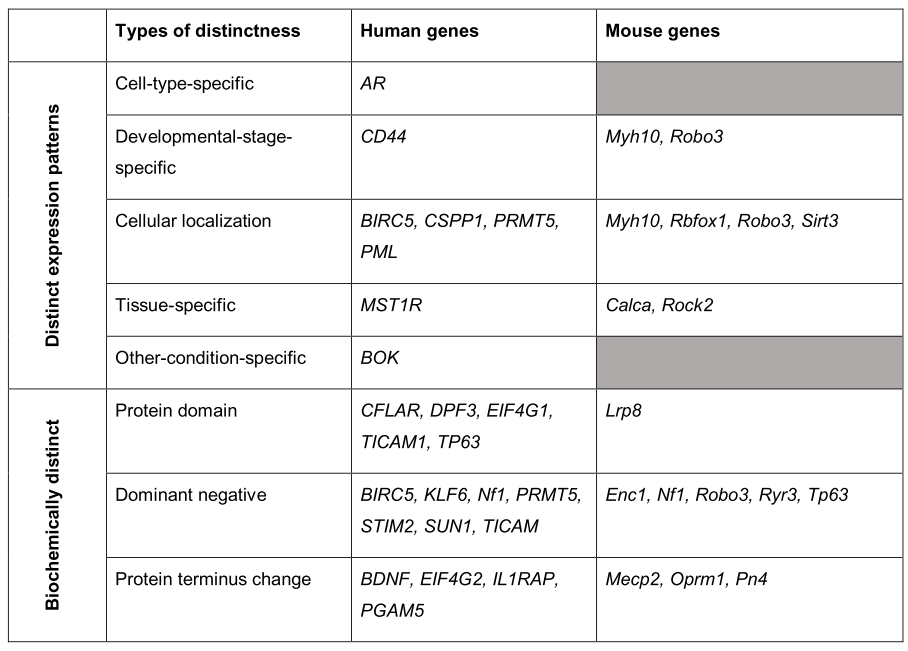
Some question
- Do most genes really have multiple functional isoforms?
- Given that only 3% of human and 9% of mouse genes have evidence for FDSIs
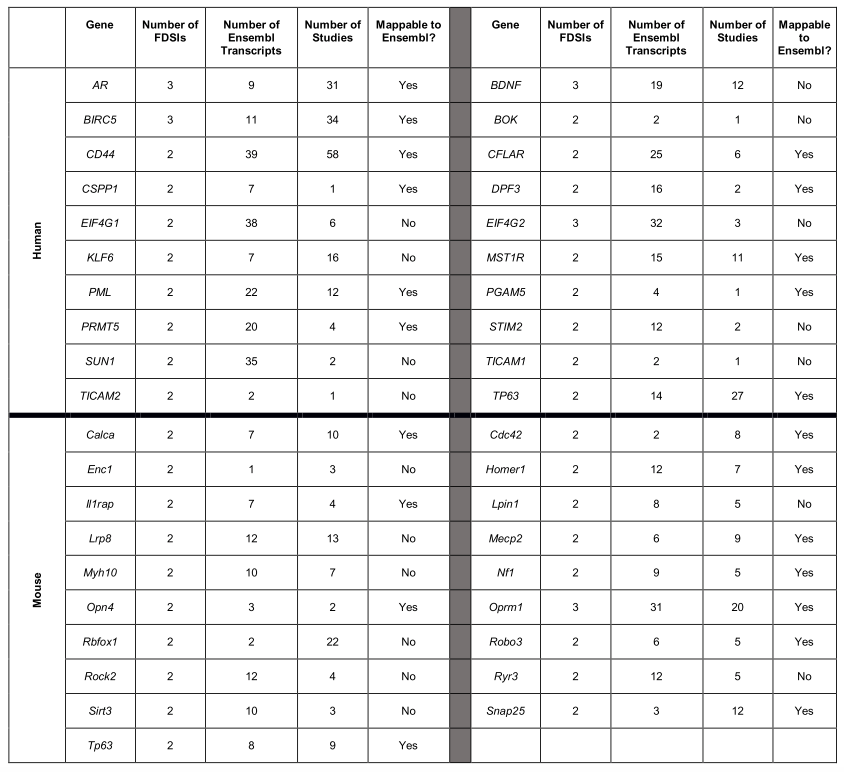
- Trying to link FDSIs back to sequence databases: 25/39
- Isoforms of 1/3 genes mentioned cannot be matched to transcripts in ENSEMBL
版权声明: 本博客所有文章除特别声明外,均采用CC BY-NC-SA 3.0 CN许可协议。转载请注明出处!